Biological Science Faculty Member
Dr. Hank W. Bass
- Office: 3074 King Life Sciences
- Office: (850) 644-9711
- Area: Cell and Molecular Biology
- Lab: King Life Sciences
- Fax: (850) 645-8447
- Mail code: 4295
- E-mail: bass@bio.fsu.edu

HW Bass Faculty Page 
Bass Lab Home Page
Professor
Ph.D., North Carolina State University, 1992.
Graduate Faculty Status
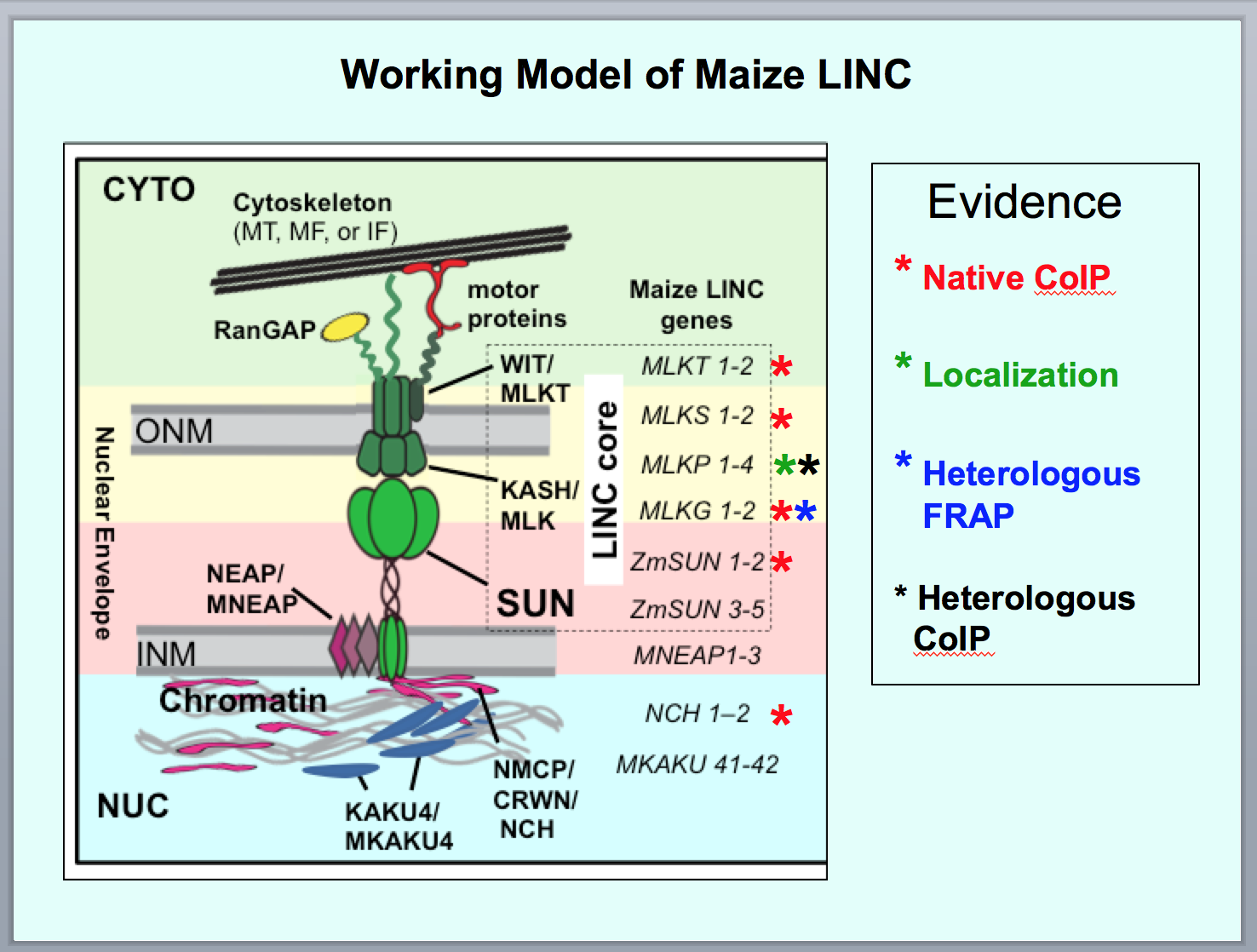
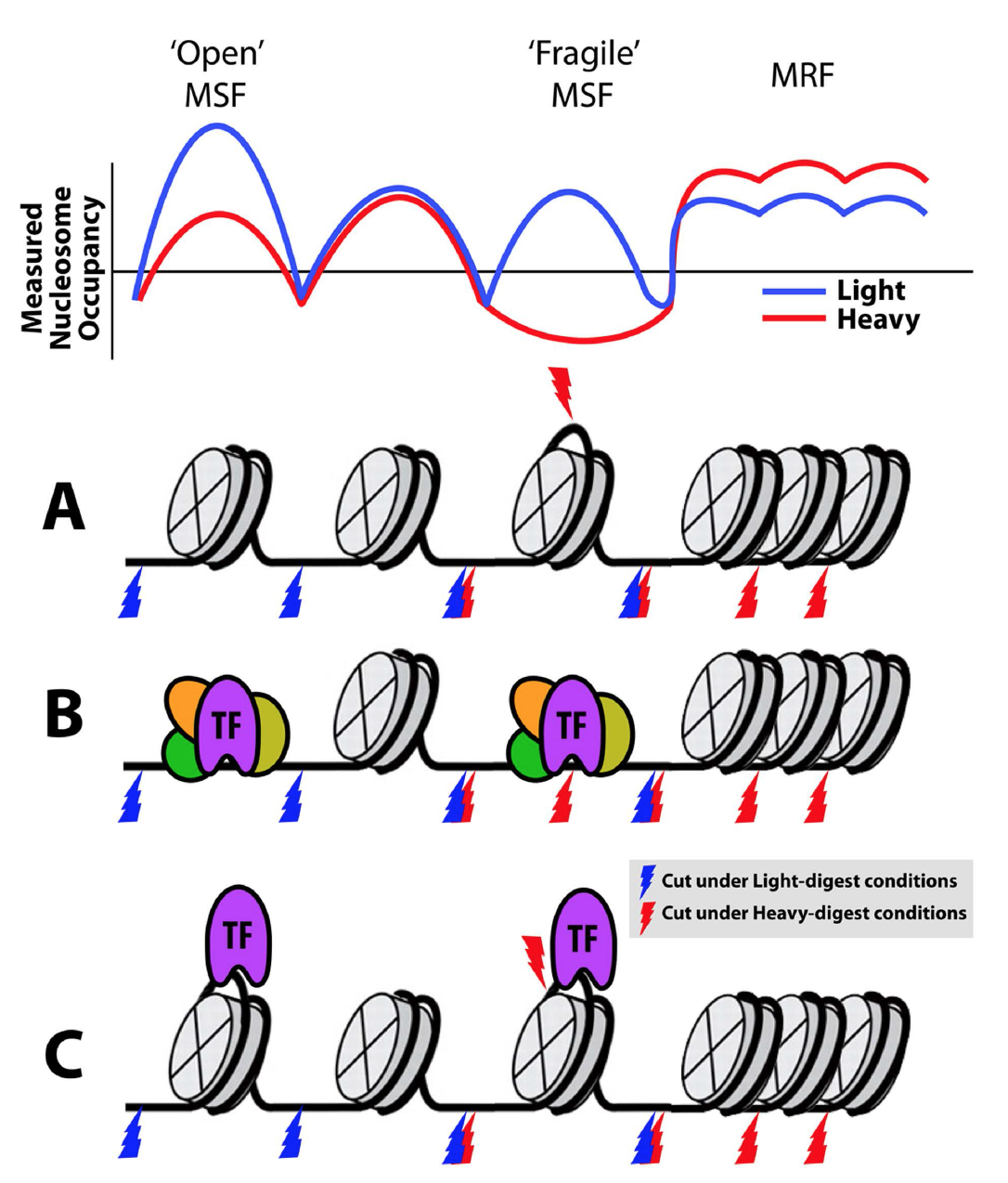
Working model of chromatin signatures from nuclease sensitivity profiling using
DNS (Differential Nuclease Sensitivity) mapping, from Vera et al., 2014.
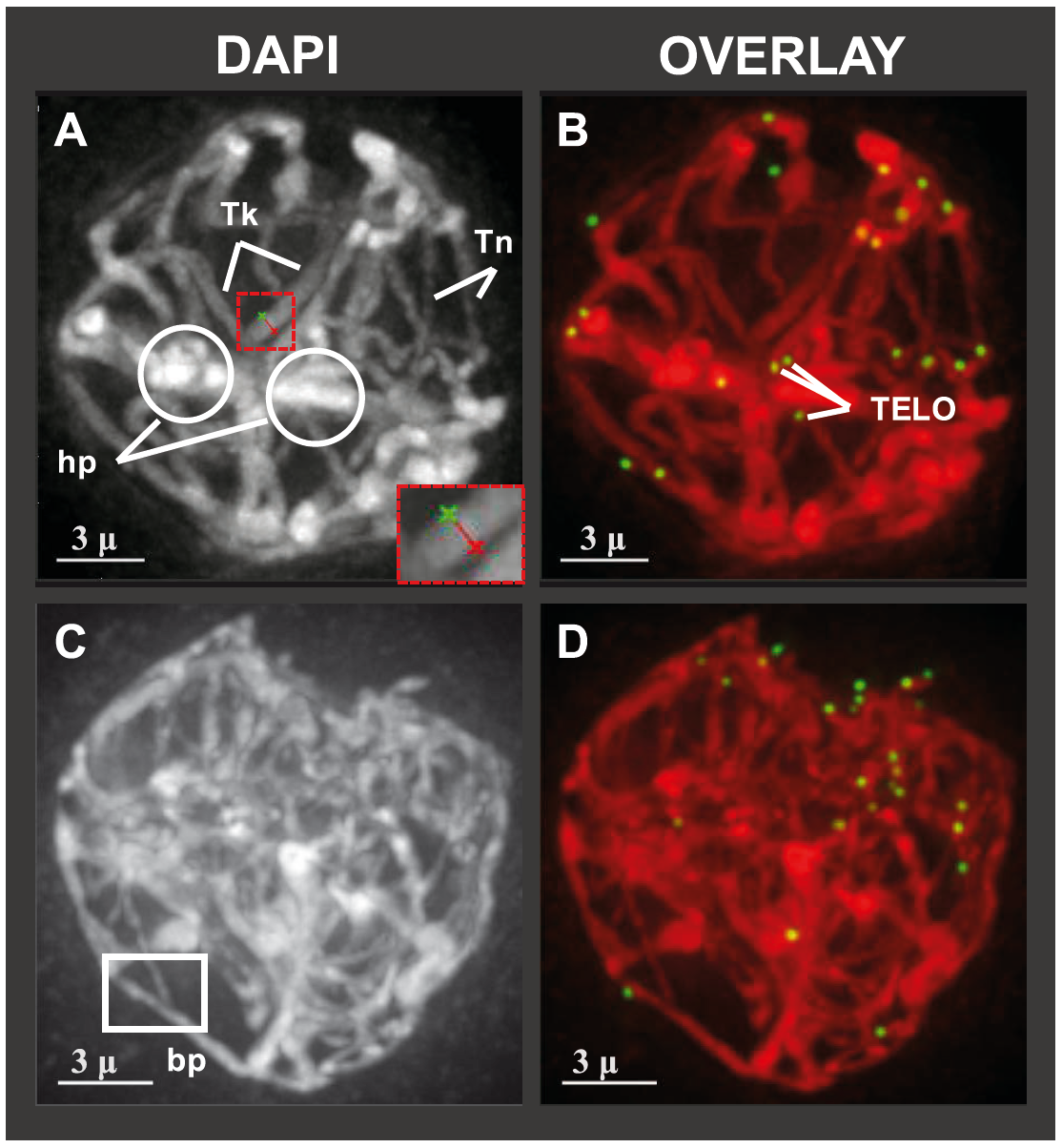
Hop meiotic prophase chromosomes and telomeres, from KA Easterling:
Research and Professional Interests:
Our work makes use of maize (corn, Zea mays L.) to define structure-function relationships of heredity and gene regulation across multiple scales. We employ genetics, microscopy, molecular biology, and genomics technologies.
Current research is advancing in several related areas, including:
Epigenomics and chromatin structure
- Spatiotemporal mapping of DNA replication in nuclei from a naturally developing multicellular organism
- MNase mapping of genome-wide chromatin sensitivity (DNS-seq) and cistrome occupany (MOA-seq) technologies for plants
- Analysis of genome-wide DNA damage signaling pathways related to hypoxiz in maize and human
Meiotic Chromosome Behavior
- Meiotic telomeres, nuclear envelope SUN-domain, LINC complex proteins in chromosome segregation
- Meiotic chromosome behavior in hop
Selected Publications:
HW Bass Publications:
on ORCID: 0000-0003-0522-0881
on Google Scholar
on Bass Lab Publications Page
Select Recent Articles:
(2024) Hu G, Grover, CE Vera DL, Lung P-Y, Girimurugan SB, Miller ER, Conover JL, Ou S, Xiong X, Zhu D, Li D, Gallagher JP, Udall JA, Sui X, Zhang J, Bass HW, and Wendel JF. "Evolutionary dynamics of chromatin structure and duplicate gene expression in diploid and allopolyploid cotton." Mol Biol Evol, Volume 41, Issue 5, May 2024, msae095. DOI: doi.org/10.1093/molbev/msae095.[PMID: 38758089].
(2023) Zhang L, Bass HW, Irianto J, and Mallory X. "Integrating SNVs and CNAs on a phylogenetic tree from single-cell DNA sequencing data." Genome Research 2023 Nov 22. doi: 10.1101/gr.277249.122. [PMID: 37993137].
(2021) Savadel* SD, Hartwig* T, Turpini ZM, Vera DL, Lung P-Y, Sui X, Blank M, Frommer WB, Dennis JH, Zhang J, and Bass HW. "The native cistrome and sequence motif families of the maize ear." (* equally contributing co-1st authors) PLOS Genetics 17(8): e1009689. DOI: 10.1371/journal.pgen.1009689.
(2021) Park M, Williams DS, Turpin ZM, Wiggins ZJ, Tsolova VM, Onokpise OU, and Bass HW. "Differential Nuclease Sensitivity Profiling Uncovers a Drought Responsive Change in Maize Leaf Chromatin Structure for two Large Retrotransposon Derivatives, Uloh and Vegu." Plant Direct 5(8), e337. DOI: doi.org/10.1002/pld3.337.
(2021) McKenna* JF, Gumber* HK, Turpin ZM, Jalovec AM, Kartick AC, Graumann K, and Bass HW. "Maize (Zea mays L.) nucleoskeletal proteins regulate nuclear envelope remodeling and function in stomatal complex development and pollen viability". (* equally contributing co-1st authors); Frontiers in Plant Science February 2021 | Volume 12 | Article 645218. doi: 10.3389/fpls.2021.645218.
(2020) Parvathaneni RK, Bertolini E, Shamimuzzaman M, Vera DL, Lung P-Y, Rice BR, Zhang J, Brown PJ, Lipka AE, Bass HW, and Eveland AL. "The regulatory landscape of early maize inflorescence development." Genome Biology 2020 Jul 6;21(1):165. doi: 10.1186/s13059-020-02070-8.
(2020) Easterling KA, Pitra NJ, Morcol TB, Aquino JR, Lopes LG, Bussey KC, Matthews PD, and Bass HW. "Identification of Tandem Repeat Families from Long-Read Sequences of Humulus lupulus." PLoS ONE 15(6): e0233971 (DOI: 10.1371/journal.pone.0233971).
(2020) Wheeler E, Brooks AM, Concia L, Vera DL, Wear EE, LeBlanc C, Ramu U, Vaughn MW, Bass HW, Martienssen RA, Thompson WF, and Hanley-Bowdoin L. "Arabidopsis DNA replication initiates in intergenic, AT-rich open chromatin." Plant Physiology 183:206-220.
(2019) Dumur T, Duncan S, Graumann K, Desset S, Randall RS, Mittelsten Scheid O, Bass HW, Prodanov D, Tatout C, adn Baroux C. "Probing the 3D architecture of the plant nucleus with microscopy approaches: challenges and solutions." Nucleus 10(1):181-212. (DOI: 10.1080/19491034.2019.1644592).
(2019) Gumber HW, McKennna JF, Tolmie AF, Jalovec AM, Kartick AC, Graumann K, and Bass HW. "MLKS2 is an ARM domain and F-actin-associated KASH protein that functions in stomatal complex development and meiotic chromosome segregation." Nucleus 10(1):144-166. (DOI: 10.1080/19491034.2019.1629795).
(2019) Gumber HK, McKenna JF, Estrada AL, Tolmie AF, Graumann K, and Bass HW. "Identification and characterization of genes encoding the nuclear envelope LINC complex in the monocot species Zea mays." Journal of Cell Science 132(3) (doi: 10.1242/jcs.221390)
(2018) Easterling KA, Pitra NJ, Jones RJ, Lopes LG, Aquino JR, Dong Z, Matthews PD, and Bass HW. "3D Molecular Cytology of Hop (Humulus lupulus) Meiotic Chromosomes Reveals Non-disomic Pairing and Segregation, Aneuploidy, and Genomic Structural Variation." Frontiers in Plant Science 9:1501(13pp) (doi /10.3389/fpls.2018.01501).
(2018) Turpin ZM, Vera DL, Savadel SD, Lung, P-Y, Wear EE, Mickelson-Young L, Thompson, WF, Hanley-Bowdoin L, Dennis, JH, Zhang J, and Bass, HW. "Chromatin Structure Profile Data from DNS-seq: Differential Nuclease Sensitivity Mapping of Four Reference Tissues of B73 Maize (Zea mays L)." Data in Brief August 2018 (DOI: 10.1016/j.dib.2018.08.015).
(2018) Springer NM, et al. "The maize W22 genome provides a foundation for functional genomics and transposon biology" Nature Genetics July 2018 (DOI: 10.1038/s41588-018-0158-0).
(2018) Girimurugan SB, Liu Y, Lung P-Y, Vera DL, Dennis JH, Bass HW, and Zhang "iSeg: an efficient algorithm for segmentation of genomic and epigenomic data." BMC Bioinformatics 19:131(15pp) (DOI: 10.1186/s12859-018-2140-3).
(2018) Griffin BD and Bass HW. "Review: Plant G-quadruplex (G4) motifs in DNA and RNA; abundant, intriguing sequences of unknown function." Plant Science 269:143-147
(DOI: 10.1016/j.plantsci.2018.01.011).
(2017) Zhang D, Easterling KA, Pitra NJ, Coles MC, Buckler ES, Bass HW, and Matthews PD. "Non-Mendelian SNP inheritance and atypical meiotic configurations are prevalent in hop (Humulus lupulusL.)" The Plant Genome 10(3):(14 pp). (DOI: 10.3835/plantgenome2017.04.0032).
(2017) Savadel SD and Bass HW. "Take a Look at Plant DNA Replication: Recent Insights and New Questions." Plant Signaling & Behavior, 12:4, e1311437.
(2016) Rodgers-Melnick E, Vera DL, Bass HW, and Buckler ES. "Open Chromatin Reveals the Functional Maize Genome." Proc Nat Acad Sci, USA 113(22):E3177-E3184.
(2015) Bass HW, Hoffman GG, Lee T-J, Wear EE, Joseph SR, Allen GC, Hanley-Bowdoin L, and Thompson WF. "Defining multiple, distinct, and shared spatiotemporal patterns of DNA replication and endoreduplication from 3D image analysis of developing maize (Zea mays L.) root tip nuclei." Plant Molecular Biology 89(4):339-351. [PubMed 26394866]
(2014) Vera DL, Madzima TF, Labonne JD, Alam MP, Hoffman GG, Girimurugan SB, Zhang J, McGinnis KM, Dennis JH, and Bass HW. "Differential nuclease sensitivity profiling of chromatin reveals biochemical footprints coupled to gene expression and functional DNA elements in maize." Plant Cell 26(10):3883-3893.
(2014) Andorf CM, Kopylov MS, Dobbs D, Koch KE, Stroupe ME, Lawrence CJ, and Bass HW. "G-quadruplex motifs in maize (Zea mays L.) occupy specific sites in thousands of genes coupled to energy status, hypoxia, low sugar, and nutrient deprivation" J Genetics Genomics 41(12):627-647.
(2014) Murphy SP, Gumber HK, Mao YY, and Bass HW. "A dynamic meiotic SUN belt includes the zygotene-stage telomere bouquet and is disrupted in chromosome segregation mutants of maize (Zea mays L.)." Frontiers in Plant Science 5:314.
(2014) Bass HW, Wear EE, Lee T-J, Hoffman GG, Gumber HK, Allen GC, Thompson WF, Hanley-Bowdoin L. "A maize root tip system to study DNA replication programmes in somatic and endocycling nuclei during plant development" J Exp Botany 65:2747-2756.
(complete list of publications page: http://fla.st/2G6ax5K)
