Biological Science Faculty Member - Retired
 Dr. Betty Jean Gaffney
Dr. Betty Jean Gaffney
| Office: | (850) 531-0693 |
| E-mail: |
bgaffney@fsu.edu |
Personal Home Page
Curriculum Vitae
These pages open in a new window. Close the window to return here.
Professor, Florida State University, 1996-2016
B.S. 1961 (Chemistry), Stanford
Ph.D. 1966 (Chemistry), Stanford
NIH Postdoctoral Fellowship 1966, Tohoku University, Sendai Japan
Varian Assoc. Postdoctoral Fellowship 1967
Research Associate, Stanford 1968-1973
NIH Research Career Development Award, 1975-80
Assistant Professor to Professor, Johns Hopkins University, 1974-1996.
Fellow of the Biophysical Society, 2009.
Fellow of the International EPR Society (IES), 2017.
Overview: Reading the biological lipid code
I have been fascinated with "reading the biological lipid code" throughout my career. That code encompasses physical properties of lipid arrays all the way to enzymes that use membrane components as substrates- different ways of rendering cell membranes dynamic. The goal is to find the messages in these dynamics. I contributed to chemical synthesis of nitroxide spin labels in the beginning and applied EPR (electron paramagnetic resonance) spectroscopy to reveal the lifetime of lipid conformations in synthetic lipid bilayers, during post-doc periods in K. Nakanishi's and in H.M. McConnell's labs [3, 4, 11].
In early studies of lipid membrane structure (1970s), it was unclear if real biological membranes and lipid model bilayers had similar properties. As a new faculty member, I showed that the abnormal appearance of transformed fibroblasts does NOT indicate a global change in lipid properties from their normal counterparts, as determined with spin labeled fatty acids [15]. In contrast, mobility of phospholipid spin labels in the close-packed membranes of Sindbis virus changes dramatically when the E1/E2 proteins are removed by proteolysis [13]. The existence of patches of membrane with specialized physical properties was foreshadowed by these early papers. I also pursued other studies of lipid dynamics, culminating in discovery (and naming) of the "sub-transition" that occurs between low temperature phases of saturated lipid bilayers [30] and in synthesis of spin-labeled, bifunctional crosslinking reagents [40, 41]. These interests broaden more recently to include the lipid remodeling enzyme lipoxygenase and new applications of biological EPR. Some results are highlighted below.
Structure and mechanism of lipoxygenases
Membrane components are constantly remodeled to signal the state of a cell and to maintain fluidity. In the early 1980s (pre-cloning), the lipid remodeling lipoxygenases were available in abundance only from soybeans and animal reticulocytes. I improved soybean lipoxygenase purification and initiated a project with crystallographers J. Boyington and L.M. Amzel. The inaugural structure of the enzyme [58, 66, 68] has become a prototype of the lipoxygenase fold (PF00305).
Two lipoxygenase types were known early on: 5- and 15-lipoxygenases, the numbers referring to the carbon bearing -OOH in products from arachidonic acid. Pathways to mediators of inflammation differ for the two products. We had solved a 15-LOX structure, and finding determinates of specificity has guided my continuing studies. Recently we determined the location and dynamics of a spin labeled lipid bound to lipoxygenase [100], using advanced pulsed EPR studies (at ACERT resource, Cornell) (Figure 1). I also contributed to structural studies by other labs interested in determinants of product stereochemistry [94], in lipoxygenase isoforms of soybean [95], in manganese lipoxygenase [88], and in a bacterial lipoxygenase-phospholipid complex [101].
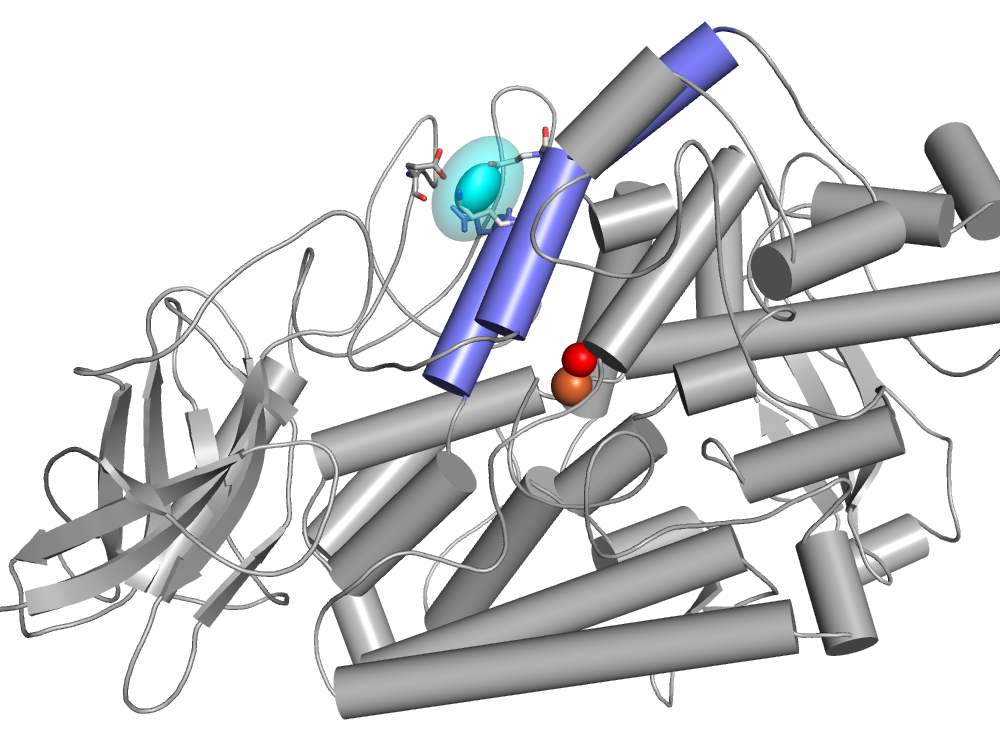
Fig. 1 Cyan ellipse: distribution of head group locations for a spin labeled lysolipid bound to soybean lipoxygenase. The lipid acyl chain must pass between the blue helices to approach the catalytic metal-water site. Iron center: rust, water: red.
The mutants of soybean lipoxygenase-1, prepared for spin label studies of structure and dynamics of the lipoxygenase [96. 100, 102], are deposited with Addgene.
Applications in Biological Magnetic Resonance
The EPR spectra of metal centers having more than one unpaired spin are intriguing and complex. I contributed to in depth reevaluation of spin 5/2 EPR spectra with quantum chemist, H. Silverstone [53]. Ferric transferrin is an historical model of non-heme iron in proteins. The mysterious EPR spectra of diferric transferrin carbonate was shown to arise from a distribution in zero field energies and the spectra could be simulated quantitatively [47]. This is confirmed in our more recent EPR measurements at the higher frequency, 94 GHz [80]. We applied similar simulation methods to studies of the non-heme iron enzymes phenylalanine hydroxylase [46] and lipoxygenases [54], showing, for instance, that only a single equivalent of lipid hydroperoxide is required to convert all the iron in resting ferrous P. aeruginosa lipoxygenase to the active ferric form [101]. We also employed higher frequency EPR as the basis for proposing that metal centers in iron and a manganese-containing lipoxygenase have similar ligand environments [88]. Further, we contributed to an NMR study in the R. Bowers lab, at UF, of hyperpolarized xenon interacting with proteins, including with lipoxygenase [84].
The complexity and beauty of metallo EPR spectra at X-band are illustrated in Figure 2, showing the many transitions occurring in a magnetic field scan of manganese lipoxygenase at 9.4 GHz [103]. Simplicity replaces beauty in a spectrum recorded at the higher frequency, 94 GHz (not shown) [88].
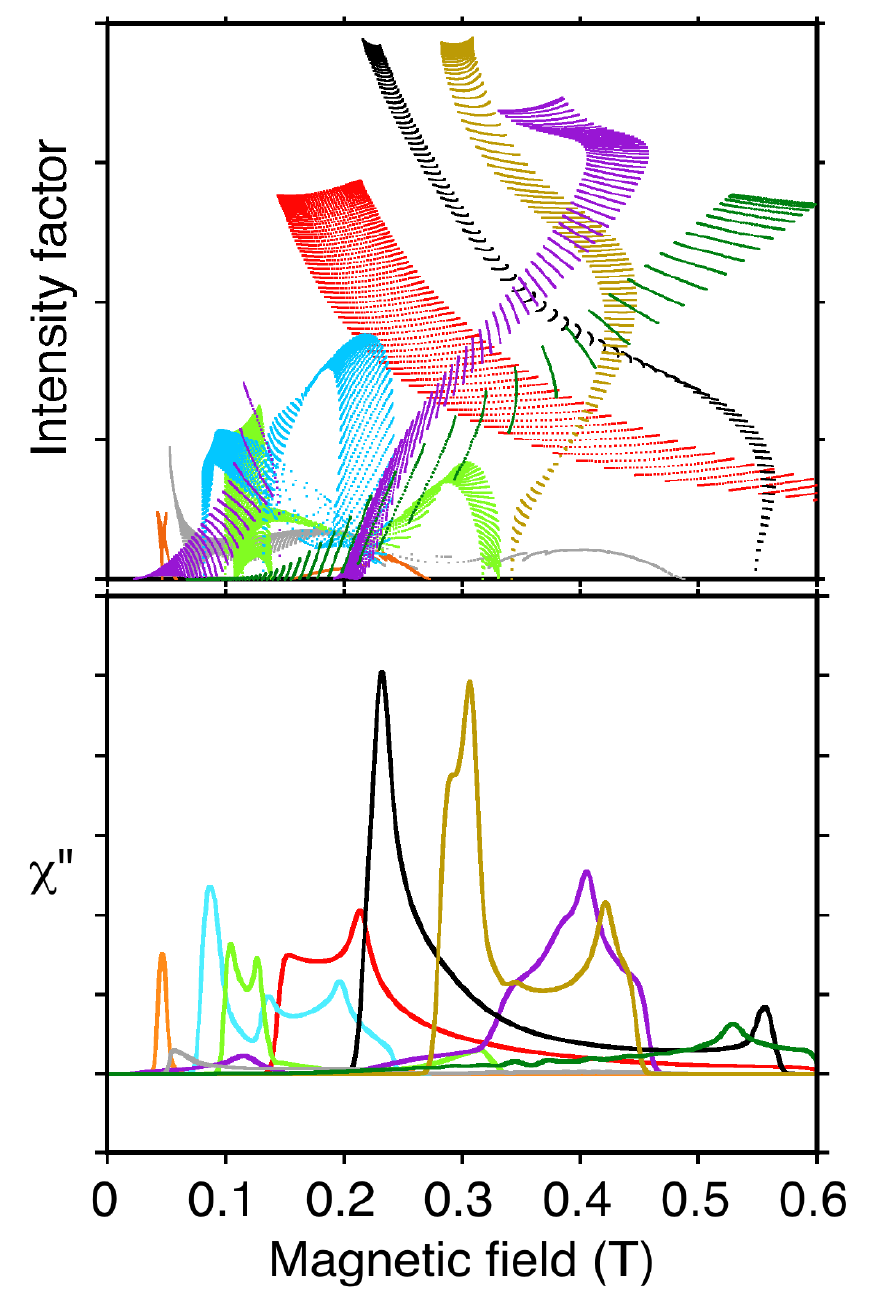
Fig. 2. Each dot (upper) represents EPR intensity from one molecular orientation in the magnetic field. These intensities sum for all orientations to give the components of observed spectra (lower). Transitions between different pairs of energy levels are represented by the various colors.
References: numbers refer to my publication list, updated in 2015, which can be found (here on my
personal home page).
For a recent review see: Betty J. Gaffney (2020) "EPR Spectroscopic Studies of Lipoxygenases", Chemistry - An Asian Journal, 15: 42-50. http://dx.doi.org/10.1002/asia.201901461.
A list of my publications can also be found at (with slightly different numbering) http://www.ncbi.nlm.nih.gov/sites/myncbi/betty.gaffney.1/bibliography/40434843/public/?sort=date&direction=descending
Selected Publications:
(Also published as Betty Gaffney McFarland, 1963-1972)
1. B. Gaffney McFarland (1963) "Ring Contraction of Steroids", in Steroid Reactions, (ed. C. Djerassi) Holden Day, Inc., San Francisco, Ch. 11, pp. 427-455.
2. B. Gaffney McFarland (1966) "Reductions by Grignard Reagents with Multiple Asymmetric Centers", Stanford University Dissertation (w. H.S. Mosher).
3. B. Gaffney McFarland, Y. Inoue and K. Nakanishi (1969) "The Reaction of Koshland's Protein Reagent with Tryptophan", Tetrahedron Letters, 10: 857-860.
4. H. M. McConnell and B. Gaffney McFarland (1970) "Physics and Chemistry and Spin Labels", Quarterly Reviews of Biophysics, 3: 91-136.
5. B. Gaffney McFarland and H. M. McConnell (1971) "Bent Fatty Acid Chains in Lecithin Bilayers", Proceedings of the National Academy of Sciences, 68: 1274-1278.
6. H. M. McConnell, K. L. Wright and B. Gaffney McFarland (1972) "The Fraction of the Lipid in a Biological Membrane that is in a Fluid State: A Spin-Label Assay", Biochemical and Biophysical Research Communications, 47: 273-281.
7. H. M. McConnell and B. Gaffney McFarland (1972) "The Flexibility Gradient in Biological Membranes", Annals of the New York Academy of Sciences, 195: 207-217.
8. B. Gaffney McFarland (1972) "The Molecular Basis of Fluidity in Membranes", Chemistry and Physics of Lipids, 8: 303-313.
9. B. J. Gaffney and C. McNamee (1974) "Spin Label Measurements in Membranes", in Biomembranes (a volume of Methods in Enzymology), Fleisher, Packer and Estabrook eds, Academic Press, N.Y., Vol. 32B, Ch. 17, pp. 161-198.
10. B. J. Gaffney and H. M. McConnell (1974) "Effect of a Magnetic Field on Phospholipid Membranes", Chemical Physics Letters, 24: 310-313.
11. B. J. Gaffney and H. M. McConnell (1974) "The Paramagnetic Resonance Spectra of Spin Labels in Phospholipid Membranes", Journal of Magnetic Resonance, 16:1-28.
12. B. J. Gaffney, P. E. Branton, G. G Wickus and C. B. Hirschberg (1974) "Fluid Lipid Regions in Normal and Rous Sarcoma Virus Transformed Chick Embryo Fibroblasts", in Viral Transformation and Endogeneous Viruses, ed. A. S. Kaplan, Academic Press, New York, pp. 97-115.
13. B.M. Sefton and B. J. Gaffney (1974) "Effect of the Viral Proteins on Fluidity of the Membrane Lipids in Sindbis Virus", Journal of Molecular Biology, 90: 343-358.
14. P. W. Robbins, G. G. Wickus, P. E. Branton, B. J. Gaffney, C. B. Hirschberg, P. Fuchs and P. M. Blumberg (1974) "The Chick Fibroblast Cell Surface after Transformation by Rous Sarcoma Virus", Cold Spring Harbor Symposium, 39: 1173-1180.
15. B. J. Gaffney (1975) "Fatty Acid Chain Flexibility in the Membranes of Normal and Transformed Fibroblasts", Proceedings of the National Academy of Sciences, 72: 664-668.
16. B.J. Gaffney (1975) "Looking at the Membrane", Cell, 4: 405-408: book reviews.
17. B. J. Gaffney (1976) "The Chemistry of Spin Labels", in Spin Labeling: Theory and Applications, ed. L. J. Berliner, Academic Press, New York, pp. 182-238.
18. B. J. Gaffney (1976) "Practical Considerations for the Calculation of Order Parameters for Fatty Acid or Phospholipid Spin Labels in Membranes", Appendix in Spin Labeling: Theory and Applications, ed. L. J. Berliner, Academic Press, New York, pp. 567-571.
19. B. J. Gaffney and D. C. Lin (1976) "Spin Label Measurements of Membrane-Bound Enzymes", in The Enzymes of Biological Membranes, ed. A. Martonosi, Plenum Press, New York, Vol. 1, Ch. 3, pp. 71-90.
20. B. J. Gaffney and R. J. Mich (1976) "A New Measurement of Surface Charge in Model and Biological Lipid Membranes", Journal of the American Chemical Society, 98: 3044-3045.
21. B. J. Gaffney and S. C. Chen (1977) "Spin Label Studies of Membranes", in Methods in Membrane Biology, ed. E. D. Korn, Plenum Press, Vol. 8, pp. 291-358.
22. B. J. Gaffney and D. C. Lin (1977) "Biophysical Techniques for Measuring Physical Properties of Lipids in Membranes", in Immunobiology of Gametes, M. Edidin and M. H. Johnson, eds., Cambridge University Press, pp. 31-40.
23. S. C. Chen and B. J. Gaffney (1978) "Paramagnetic Resonance Evidence for Phase Transitions in Bilayers of Pure Spin Labeled Lipids", Journal of Magnetic Resonance, 29: 341-353.
24. C. L. Wey, R. A. Cone and B. J. Gaffney (1979) "Effects of Light and C-GMP on Membrane Fluidity of Frog Rod Outer Segments", Photochemistry and Photobiology, 29: 707-713.
25. C.L. Wey, P.L. Ahl, R.A. Cone,and B.J. Gaffney (1979) "Membrane Viscosity: A Small Lipid-Phase Spin Probe Reports the Same Viscosity as Two Integral Membrane Proteins", Biophys. J., 25: 169a.
26. C.L. Wey, R.A. Cone and B.J. Gaffney (1979) "Diffusion in Membranes: How much does the Viscosity of the Aqueous Phase Affect Diffusion Rates of Membrane Proteins and Lipids?", Biophys. J., 25: 284a.
27. B. M. Sefton and B. J. Gaffney (1979) "Complete Exchange of Viral Cholesterol", Biochemistry, 18: 436-442.
28. B. J. Gaffney (1979) "Spin Label-Thiourea Adducts: A Model for Saturation Transfer EPR Studies of Slow, Anisotropic Rotation", The Journal of Physical Chemistry, 83: 3345-3349.
29. B. J. Gaffney, D. B. Drachman, D. C. Lin and G. Tennekoon (1980) "Spin-Label Studies of Erythrocytes in Myotonic Dystrophy: No Increase in Membrane Fluidity", Journal of Neurology, 30: 272-276.
30. S. C. Chen, J. M. Sturtevant and B. J. Gaffney (1980) "Scanning Calorimetric Evidence for a Third Phase Transition in Phosphatidylcholine Bilayers", Proceedings of the National Academy of Sciences, 77: 5060-5063.
31. B. J. Gaffney, J. P. A. Scibilia and C. Elbrecht (1980) "Conventional EPR of 15N Nitroxides is Sensitive to Correlation Times as Long as 10-4 Sec", Bulletin of Magnetic Resonance, p. 374.
32. M. W. Tse-Tang, B. J. Gaffney and R. E. Kelly (1981) "Synthesis of Bifunctional Spin Label Molecules and Their Orientations in Membranes", Heterocycles, 15: 965-974.
33. B. J. Gaffney, C. H. Elbrecht and J. P. A. Scibilia (1981) "Enhanced Sensitivity to Slow Motions Using 15N-Nitroxide Spin Labels", J. Magn. Res., 44: 436-446.
34. R. K. Scheule and B. J. Gaffney (1981) "Physical Properties of Native and Reconstituted Membranes Containing Sindbis Virus Glycoproteins, in Liposomes, Drugs and Immunocompetent Cell Functions, ed. C. Nicolau and A. Paraf, Academic Press, London, pp. 79-93.
35. R. K. Scheule and B. J. Gaffney (1981) "Reconstitution of Membranes with Fractions of Triton X-100 Which are Easily Removed", Anal. Biochem., 117: 61-66.
36. B. J. Gaffney and R. K. Scheule (1982) "What is a Successful Reconstitution of a Membrane Glycoprotein that Lacks an Enzymatic Activity?", Biophys. J., 37: 81-82 (Discussions book).
37. S.-C. Chen, J. M. Sturtevant, K. Conklin and B.J. Gaffney (1982) "Calorimetric Evidence for Phase Transitions in Spin-Labeled Lipid Bilayers", Biochemistry, 21: 5096-5101.
38. H. Grill, N. Weigel, B. J. Gaffney and S. Roseman (1982) "Sugar Transport by the Bacterial Phosphotransferase System: XVII. Radioactive and Electron Paramagnetic Resonance Labeling of the Salmonella typhimurium Phosphocarrier Protein HPr at the Amino-terminal Methionine", J. Biol. Chem., 257: 14510-14517.
39. R. A. Lazarus, D. E. Wallick, R. F. Dietrich, D. W. Gottschall, S. J. Benkovic, B. J. Gaffney and R. Shiman (1982) "The Mechanism of Phenylalanine Hydroxylase", Federation Proceedings, 41: 2605-2607.
40. B. J. Gaffney, G. L. Willingham and R. S. Schepp (1983) "Synthesis and Membrane Interactions of Spin Label Bifunctional Reagents", Biochemistry, 22: 881-892.
41. G. L. Willingham and B. J. Gaffney (1983) "Reactions of Spin Label Crosslinking Reagents with Red Blood Cell Proteins", Biochemistry, 22: 892-898.
42. D. E. Wallick, L. M. Bloom, B. J. Gaffney and S. J. Benkovic (1984) "The Reductive Activation of Phenylalanine Hydroxylase and Its Effect on the Redox State of the Nonheme Iron", Biochemistry, 23: 1295-1302.
43. S. Benkovic, D. Wallick, L. Bloom, B. J. Gaffney, P. Domanico, T. Dix and S. Pember (1985) "On the Mechanism of Action of Phenylalanine Hydroxylase", Biochem. Soc. Transactions, 13: 436-438.
44. B. J. Gaffney (1985) "Chemical and Biochemical Crosslinking of Membrane Components", Biochim. Biophys. Acta Reviews on Biomembranes, 822: 289-317.
45. S. J. Benkovic, L. M. Bloom, G. Bollag, T. A. Dix, B. J. Gaffney and S. Pember (1986) "The Mechanism of Action of Phenylalanine Hydroxylase", Ann. N.Y. Acad. Sci., 471: 226-232.
46. L. M. Bloom, S. J. Benkovic and B.J. Gaffney (1986) "Characterization of Phenylalanine Hydroxylase", Biochemistry, 25: 4204-4210.
47. A.-S. Yang and B. J. Gaffney (1987) "Determination of Relative Spin Concentration in Some High-Spin Ferric Proteins Using E/D-Distribution in Electron Paramagnetic Resonance Simulations", Biophys. J., 51: 55-67.
48. J. C. Boyington, B. J. Gaffney and L. M. Amzel (1990) "Crystallization and Preliminary X-ray Analysis of Soybean Lipoxygenase-1, A Non-heme Iron-containing Dioxygenase", J. Biol. Chem., 265: 12771-12773.
49. A.-S. Yang and B.J. Gaffney (1990) "EPR Detection of Kinetic Responses to Photochemically Generated Protein Cofactors", J. Magn. Res., 90: 580-583.
50. B.J. Gaffney (1990), book review of "Spin Labelling: Theory and Applications. Biological Magnetic Resonance. Volume 8", J. Am. Chem. Soc., 112: 6750.
51. J. Dubach, B.J. Gaffney, K. More, G.R. Eaton, and S.S. Eaton (1991) "The Effect of the Synergistic Anion on EPR Spectra of Iron-Transferrin Anion Complexes is Consistent with Bidentate Binding of the Anion", Biophys. J., 59: 1091-1100.
52. B.J. Gaffney and P. J. Dagdigian (1993) "Oxygen Binding to Hemoglobin", in Physical Chemistry, Developing a Dynamic Curriculum, R.W. Schwenz and R.J. Moore eds., American Chemical Society, Ch. 25 , pp. 370-379.
53. B. J. Gaffney and H. J. Silverstone (1993) "Simulation of the EMR Spectra of High Spin Iron in Proteins", a Chapter in Biological Magnetic Resonance, Vol. 13: EMR of Paramagnetic Molecules, L.J. Berliner and J. Reuben, Eds, Plenum N.Y., pp 1-57.
54. B. J. Gaffney, D. V. Mavrophilipos and K.S. Doctor (1993) "Access of Ligands to the Ferric Center in Lipoxygenase-1", Biophys. J., 64: 773-783.
55. K. S. Doctor, B. J. Gaffney, G. Alvarez and H. J. Silverstone (1993) "EPR Spectroscopy of Interdoublet Transitions in High-Spin Iron: Applications to Transferrin Oxalate", J. Phys. Chem., 97: 3028-3033.
56. D.D. Thomas. B.J. Gaffney and O.H. Griffith (1993) "The Science Speaks for Itself: a Biographical Sketch of Harden McConnell", Biophys. J., 64: 573-576.
57. O.H. Griffith, B.J. Gaffney and A.L. Kwiram (1993) "Harden M. McConnell: A Celebration of His Scientific Achievements", J. Phys. Chem., 97: 2807-2810.
58. J.C. Boyington, B. J. Gaffney, and L.M. Amzel (1993) "The Three-Dimensional Structure of an Arachidonic Acid 15-Lipoxygenase", Science, 260: 1482-1486.
59. J.C. Boyington, B. J. Gaffney, and L.M. Amzel (1993) "Structure of Soybean Lipoxygenase-1", Biochem. Soc. Transactions, 21: 744-748.
60. J.C. Boyington, B. J. Gaffney, and L.M. Amzel (1994) "The Three-Dimensional Structure of Soybean Lipoxygenase-1: an Arachidonic Acid 15-Lipoxygenase", in "Eicosanoids and Other Bioactive Lipids in Cancer, Inflammation and Radiation Injury", K.V. Honn, S. Nigam and L.J. Marnett, eds., Kluwer Acad. Pub., Norwell, MA, pp.131-136.
61. J.C. Boyington, B. J. Gaffney, L.M. Amzel, K.S. Doctor, D.V. Mavrophilipos, Z.V. Mavrophilipos, A.Colom and S.M. Yuan (1994) "The X-ray Structure and Biophysical Studies of a 15-Lipoxygenase" Annals. New York Acad. Sci., 744: 310-313. Figure on cover.
62. B.J. Gaffney and B.C. Maguire (1995) "Nuclear Magnetic Resonance of Biomolecules in Solution" for Encyclopedia of Molecular Biology and Biotechnology, R.A. Meyers ed., VCH Publishers, New York, pp. 601-604.
63. B.J. Gaffney, J.C. Boyington, L.M. Amzel, K.S. Doctor, S.T. Prigge, S.M. Yuan (1995) "Lipoxygenase Structure and Mechanism" Proc. 9th Int. Conf. on Prostaglandins and Related Compounds, in "Advances in Prostaglandin, Thromboxane and Leukotriene Research", v. 23, Raven Press, New York, pp. 11-16.
64. M.W. Crowder, J.D. Stewart, V.A. Roberts, C.J. Bender, E Tevelrakh, S.D. Taylor, P.C.D. Hawkins, S. Franklin, J. Peisach, E.D. Getzoff, B.J. Gaffney and S. J. Benkovic (1995) "Spectroscopic Studies on the Designed Metal-Binding Sites of the 43C9 Single Chain Antibody", J. Am. Chem. Soc., 117: 5627-5634.
65. L.M. Amzel and B.J. Gaffney (1995) "Structural Immunology: Problems in Molecular Recognition", FASEB J., 9: 7-8.
66. S.T. Prigge, J.C. Boyington, B.J. Gaffney and L.M. Amzel (1996) "Structure Conservation in Lipoxygenases: Structural Analysis of Soybean Lipoxygenase-1 and Modeling of Human Lipoxygenases", Proteins: Structure, Function, and Genetics, 24: 275-291.
67. B.C. Maguire and B.J. Gaffney (1996) "Nuclear Magnetic Resonance of Biological Molecules in Solution" in Encyclopedia of Molecular Biology , Fundamentals and Applications, VCH Publishers, New York, pp. 218-226.
68. B.J. Gaffney (1996) "Lipoxygenases: Structural Principles and Spectroscopy" Ann. Reviews Biophys. Biomol. Struct., 25:.431-59.
69. S.T. Prigge, J.C. Boyington, B.J. Gaffney and L.M. Amzel (1996) "Lipoxygenase: Structure and Function", in "Lipoxygenases and Lipoxygenase Pathway Enzymes", G. Piazza ed. AOCS Press, Champaign, IL, Ch. 1, pp. 1-32.
70. K.S. Doctor and B.J. Gaffney (1996) "High Frequency EPR Predictions for the Non-heme Iron Protein Lipoxygenase" Appl. Magn. Res., 11: 425-435.
71. B.J. Gaffney (1996) "Origins of Biological Magnetic Resonance" FASEB J. 10: 1448-1451.
72. B. C. Maguire and B. J. Gaffney (1997) "Interdoublet Transitions in S=5/2 Protein Systems" Solid State NMR, 9: 81-83.
73. J.C. Boyington, B. J. Gaffney and L.M. Amzel (1997) "The Three Dimensional Structure of Soybean Lipoxygenase-1: An Arachidonic Acid 15-Lipoxygenase" Adv. Exp. Med. Biol. 400A: 133-138.
74. S.T. Prigge, J.C. Boyington, M. Faig, K.S. Doctor, B.J. Gaffney and L.M. Amzel (1997) "Structure and Mechanism of Lipoxygenases" Biochimie, 79: 629-636.
75. B.J. Gaffney (1998) "Biochemical Spectroscopy and Dynamics" in Principles of Chemistry in Biology, E.C. Theil ed., American Chemical Society, Washington, D.C., Ch 6, pp. 157-186.
76. S.T. Prigge, B.J. Gaffney and L.M. Amzel (1998) "Relation Between Positional Specificity and Chirality in Mammalian Lipoxygenases" Nature Structural Biology, 5: 178-9.
77. B.J. Gaffney and H.J. Silverstone (1998) "Simulation Methods for Looping Transitions", J. Magn. Res., 134: 57-66. Figure on cover.
78. B.J. Gaffney, G.R. Eaton and S.S. Eaton (1998) "Electron Spin Relaxation Rates for High-spin Fe(III) in Iron Transferrin Carbonate and Iron Transferrin Oxalate" J. Phys Chem. 102: 5536-5541.
79. B.J. Gaffney and D. Marsh (1998) "High Frequency, Spin Label EPR of Non-Axial Ordering and Motion in Cholesterol-containing Membranes" Proc. Natl. Acad. Sci. U.S. 95: 12940-12943.
80. B.J. Gaffney, B.C. Maguire, R.T. Weber and G.G. Maresch (1999) "Disorder at Metal Sites in Proteins: a High Frequency EMR Study" Appl. Magn. Res., 16: 207-222.
81. B.J. Gaffney "Electron Magnetic Resonance" (1999) Encyclopedia of Molecular Biology, T.E. Creighton ed., John Wiley, New York, 792-795
82. B.J. Gaffney "Spin Labeling" (1999) Encyclopedia of Molecular Biology, T.E. Creighton ed., John Wiley, New York, 2408-9.
83. D. Marsh, V. A. Livshits, T. Páli and B. J. Gaffney (1999) "Recent developments in biological spin-label spectroscopy" Spectroscopy of Biological Molecules: New Directions J. Greve, G.P. Puppels and C. Otto eds., Kluwer, Dordrecht, pp. 647-650.
84. C.R. Bowers, V. Storhaug, Charles Edwin Webster, J. Bharatam, A. Cottone, III, R. Gianna, K. Betsey and B.J. Gaffney (1999) "Exploring Protein Surfaces and Cavities in Lipoxygenase and other Proteins by Hyperpolarized Xenon-129 NMR" J. Am. Chem. Soc., 121: 9370-9377.
85. C.V. Grant, W. Cope, J.A. Ball, K. Campbell, G.G. Maresch, B.J. Gaffney, W. Fink and R.D. Britt (1999) "Electronic Structure of the Aqueous Vanadyl Ion Probed by 9 and 94 GHz EPR and Pulsed ENDOR Spectroscopies and Density Functional Theory Calculations" J. Phys. Chem. B. 103: 10627-10631.
86. B.J. Gaffney and H.J. Silverstone (1999) "anisoironhs.f and cubspegiron.f: FORTRAN programs for S=5/2 EMR Spectra with Distributions in Zero field Splittings" in Cammack, R. and Fann, Y. eds. (1995-1999) EPR Software database: a compendium of programs from and for EPR spectroscopists; listed by the Intl. EPRSociety. (http://epr.niehs.nih.gov/).
87. B. Abraham, M. Sono, O. Boutaud, A. Shriner, J. H. Dawson, A. R. Brash and B. J. Gaffney (2001) "Characterization of the Coral Allene Oxide Synthase Active Site with Magnetic Circular Dichroism and Electron Paramagnetic Resonance Spectroscopy: Evidence for Tyrosinate-Ligation to the Ferric Enzyme Heme Iron" Biochemistry, 40: 2251-2259.
88. B.J. Gaffney, C. Su and E.H. Oliw (2001) "Assignment of EPR Transitions in a Manganese-Containing Lipoxygenase and Prediction of Local Structure" Appl. Magn. Res., 21: 411-422; minor correction (2004) 26: 457.
89. B.J. Gaffney (2002) "Electron Paramagnetic Resonance" Encyclopedia of Molecular Biology, T.E. Creighton ed., Wiley Online Library, New York, (online update of pub. # 81). doi: 10.1002/047120918X.emb0460
90. B.J. Gaffney (2002) "Spin Labeling" Encyclopedia of Molecular Biology, T.E. Creighton ed., Wiley Online Library, New York, (online update of # 82). doi:10.1002/047120918X.emb1456.
91. F. Wu, L.J. Katsir, M. Seavy and B.J. Gaffney (2003) "Role of Radical Formation at Y193 in the Allene Oxide Synthase Domain of a Lipoxygenase-AOS Fusion Protein from Coral" Biochemistry, 42: 6871-6880.
92. B.J. Gaffney (2004) "Electron Spin Resonance of Biomolecules" Encyclopedia of Molecular Cell Biology and Molecular Medicine, Vol. 4, R.A. Meyers ed. Wiley-VCH, Weinheim, Germany, 115-133.
93. S. Agarwalla, R.M. Stroud, B.J. Gaffney (2004) "Redox Reactions of the Iron-Sulfur Cluster in a Ribosomal RNA Methyltransferase, RumA: Optical and EPR Studies" J. Biol. Chem., 279: 34123-34129.
94. G. Coffa, A.N. Imber, B.C. Maguire, G. Laxmikanthan, C. Schneider, B.J. Gaffney, and A.R. Brash (2005) "On the Relationships of Substrate Orientation, Hydrogen Abstraction and Product Stereochemistry in Single and Double Dioxygenations by Soybean Lipoxygenase-1 and Its Ala542Gly Mutant" J. Biol. Chem. 280: 38756-38766./span>
95. B. Youn, G.E. Sellhorn, R.J. Mirchel, B.J. Gaffney, H.D. Grimes, C.H. Kang (2006) "Crystal Structures of Vegetative Soybean Lipoxygenase VLX-B and VLX-D, and comparisons with seed isoforms, LOX-1 and LOX-3" PROTEINS: Structure, Function, and Bioinformatics 65: 1008-1020.
96. F. Wu and B.J. Gaffney (2006) "Dynamic Behavior of Fatty Acid Spin Labels within a Binding Site of Soybean Lipoxygenase-1" Biochemistry 45: 12510-12518.
97. B.J. Gaffney (2006) "Notes on postdocing with Harden McConnell" EPR newsletter, available online at http://hardenmcconnell.org/addendum_II_Gordon_Conference.html,vol.15 no.4, pp. 10-12.
98. B.J. Gaffney (2007) "Anesthesia, Analgesia and Euphoria", Biophys. J. 92: 1-2.
99. B.J. Gaffney (2009) "High Resolution EPR of Mononuclear Iron Proteins", Chapter 6 in Biological Magnetic Resonance, Vol 28. High Resolution EPR: Applications to Metalloenzymes and Metals in Medicine, G. Hanson and L.J. Berliner eds., Springer Pub., pp. 233-268. DOI: 10.1007/978-0-387-84856-3_6. PMC2860145.
100. B.J. Gaffney, M.D. Bradshaw, S. Frausto, F. Wu, J.H. Freed and P.Borbat. (2012) "Locating a lipid at the portal to the lipoxygenase active site" Biophys. J., 103: 2134-2144. PMC3512035. With long supplement:http://www.biophysj.org/biophysj/supplemental/S0006-3495(12)01105-8
101. A. Garreta, S. Val-Moraes, Q. García-Fernández, M. Busquets, C. Juan, A. Oliver, A. Ortiz, B.J. Gaffney, I. Fita, À. Manresa and X. Carpena (2013) "Structure and Interaction with Phospholipids of a Prokaryotic Lipoxygenase from Pseudomonas aeruginosa", FASEB J, 27: 4811-4821, doi: 10.1096/fj.13-235952; PMID: 23985801.
102. M.D. Bradshaw and B.J. Gaffney (2014) "Fluctuations of an exposed pi-helix involved in lipoxygenase substrate recognition" Biochemistry, 53: 5102-5110. doi: 10.1021/bi500768c; PMC4131896.
103. B.J. Gaffney (2014) "Connecting Lipoxygenase Function to Structure by EPR" Accounts of Chemical Research, 47: 3588-3595. http://dx.doi.org/10.1021/ar500290r.
104. Betty J. Gaffney (2020) "EPR Spectroscopic Studies of Lipoxygenases", Chemistry - An Asian Journal, 15: 42-50. http://dx.doi.org/10.1002/asia.201901461

 :
External sites will open in a new browser window.
:
External sites will open in a new browser window.