Computational Biology Program
Biology Track
Biology Track
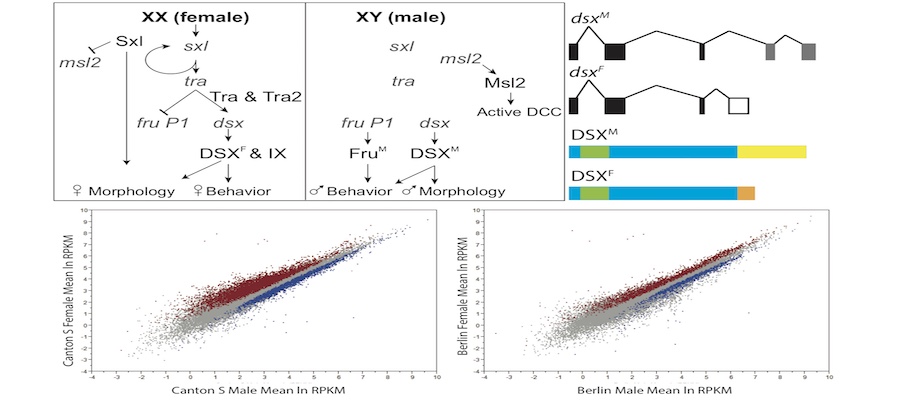 The Arbeitman lab studies the molecular-genetic basis of complex behavior using Drosophila melanogaster as a model. The Drosophila model system has some of the most sophisticated tools to understand behavior with cell-type resolution, given that the neurons that underlie complicated reproductive behaviors have been identified using molecular tools. Students can participate on computational studies of genomic data sets that were generated to understand complex reproductive behaviors. The student should have familiarity with genetic and molecular biology concepts and should be able to work independently using python and R programming tools.
The Arbeitman lab studies the molecular-genetic basis of complex behavior using Drosophila melanogaster as a model. The Drosophila model system has some of the most sophisticated tools to understand behavior with cell-type resolution, given that the neurons that underlie complicated reproductive behaviors have been identified using molecular tools. Students can participate on computational studies of genomic data sets that were generated to understand complex reproductive behaviors. The student should have familiarity with genetic and molecular biology concepts and should be able to work independently using python and R programming tools.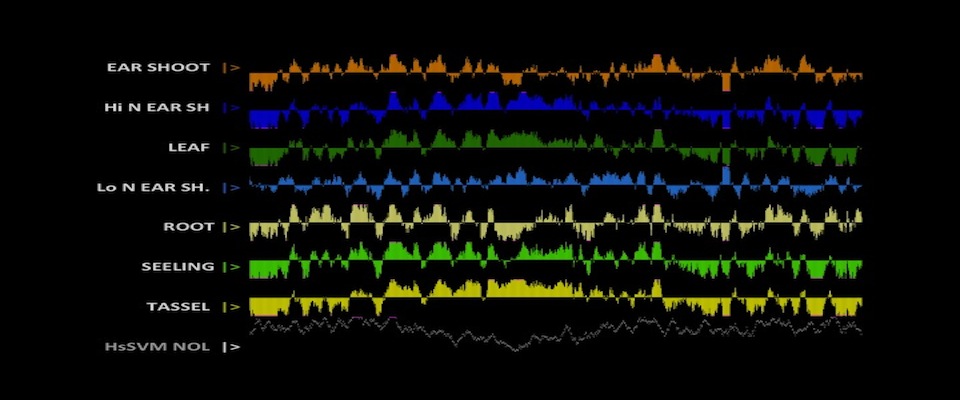 Analysis of maize genomic and epigenomic data in maize. Ongoing projects include chromatin structure profiling, analysis of genomic G-quadruplex motifs, or computational analysis of 3D images of nuclei. Student should be fluent in unix, file manipulation, and adept at self-teaching from online documentation of commonly used bioinformatic tools. Student should also have good organizational and note-taking skills along with a basic knowledge of genetics and biology.
Analysis of maize genomic and epigenomic data in maize. Ongoing projects include chromatin structure profiling, analysis of genomic G-quadruplex motifs, or computational analysis of 3D images of nuclei. Student should be fluent in unix, file manipulation, and adept at self-teaching from online documentation of commonly used bioinformatic tools. Student should also have good organizational and note-taking skills along with a basic knowledge of genetics and biology.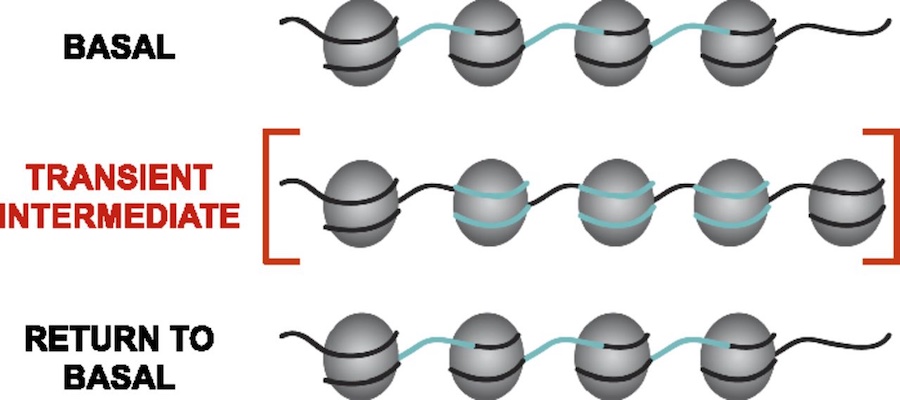 The objective of work in the Dennis lab is to characterize the changes in nucleosome organization in development and disease. We have shown that chromatin regulatory proteins control widespread and transient remodeling events that result in altered nucleosome sensitivities that define the potential of the cell. Internships in the Dennis lab will use computational approaches to facilitate an integrated analysis of dynamic nucleosome remodeling and nucleosome sensitivity. These studies will yield immediate and widely applicable information on the fundamental biology of complex genomes and ultimately new approaches for understanding development and disease.
The objective of work in the Dennis lab is to characterize the changes in nucleosome organization in development and disease. We have shown that chromatin regulatory proteins control widespread and transient remodeling events that result in altered nucleosome sensitivities that define the potential of the cell. Internships in the Dennis lab will use computational approaches to facilitate an integrated analysis of dynamic nucleosome remodeling and nucleosome sensitivity. These studies will yield immediate and widely applicable information on the fundamental biology of complex genomes and ultimately new approaches for understanding development and disease. Epigenetic regulation has recently emerged as an underpinning mechanim of brain disorders, particularly neuropsychiatric diseases. We will utilize cutting edge epi/genomic tools to decipher out novel epigenetic mechanims with an interdesciplinary intergration with behavioral neuroscience. We are particularly interested in the roles of non-coding RNAs and newly defined DNA epigenetic modifications in drug addiction and depression by using mouse models. A basic knowledge of biology and genetics and skills of computation programing are required. You will have an opportunity to work on the next generation sequencing data analyses (DNA methylation seq, mRNAseq, small RNAseq, ChIPseq) within and beyond the laboratory.
Epigenetic regulation has recently emerged as an underpinning mechanim of brain disorders, particularly neuropsychiatric diseases. We will utilize cutting edge epi/genomic tools to decipher out novel epigenetic mechanims with an interdesciplinary intergration with behavioral neuroscience. We are particularly interested in the roles of non-coding RNAs and newly defined DNA epigenetic modifications in drug addiction and depression by using mouse models. A basic knowledge of biology and genetics and skills of computation programing are required. You will have an opportunity to work on the next generation sequencing data analyses (DNA methylation seq, mRNAseq, small RNAseq, ChIPseq) within and beyond the laboratory.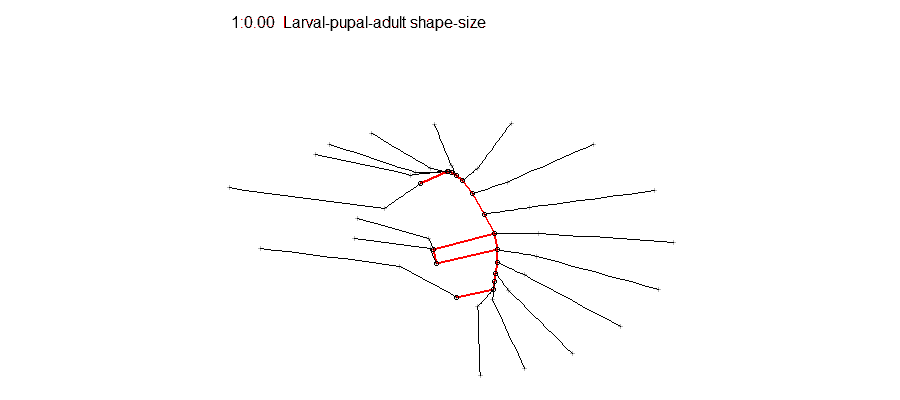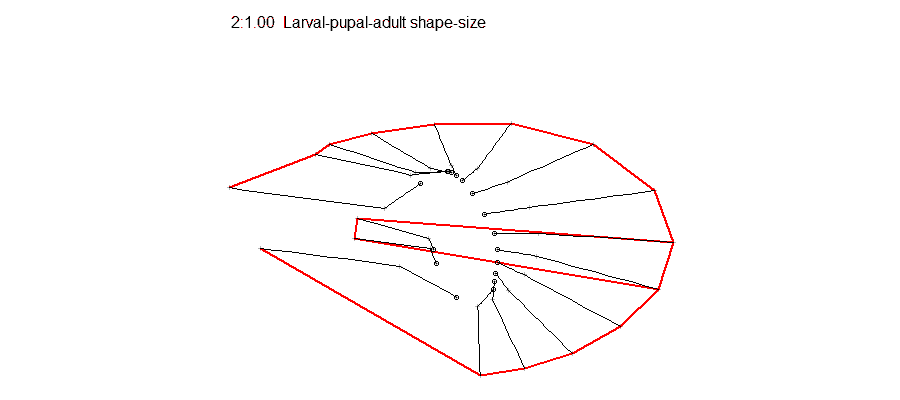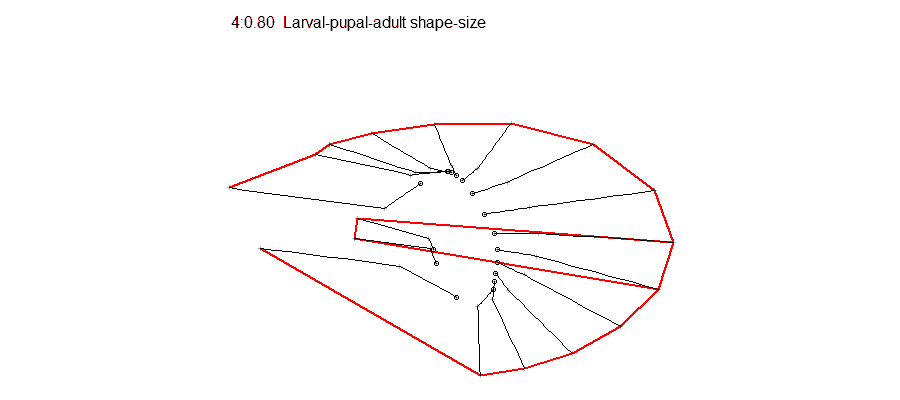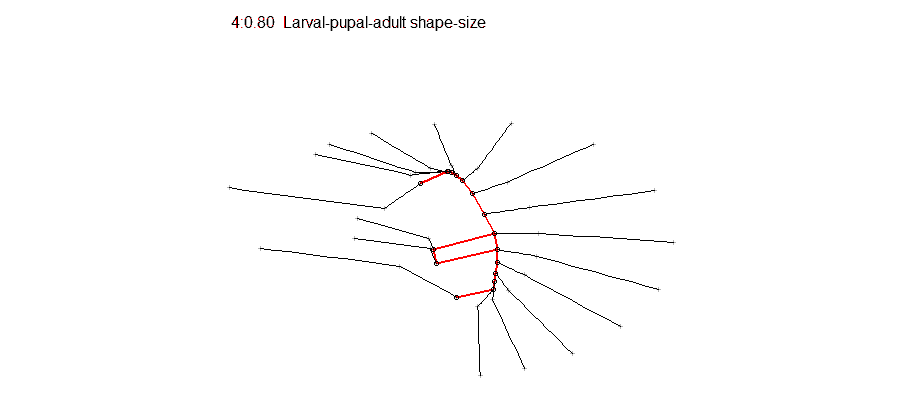 Opportunities for students to work on modeling, simulation, visualization or data analysis on genomic and phenotypic data on the relationship between development and evolution in the fruit fly, Drosophila melanogaster. We can take advantage of a variety of your skills, including programming, bioinformatics or statistics. Students should have background in genetics and preferably evolutionary biology.
Opportunities for students to work on modeling, simulation, visualization or data analysis on genomic and phenotypic data on the relationship between development and evolution in the fruit fly, Drosophila melanogaster. We can take advantage of a variety of your skills, including programming, bioinformatics or statistics. Students should have background in genetics and preferably evolutionary biology.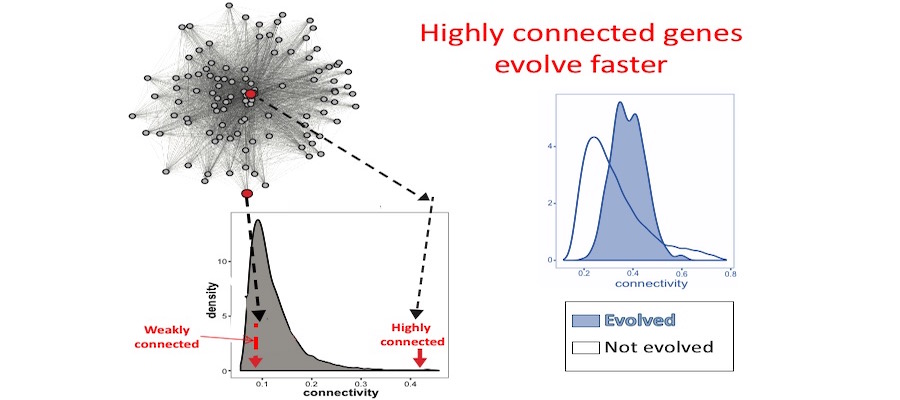 Analyze genetic and genomic data that addresses fundamental questions such as, “why are individuals within species so genetically diverse, and what genetic, ecological and evolutionary processes promote that diversity?” Student is expected to have experience with linux command line and shell scripting, at least one coding language such as C/C++, Python, or R, basic knowledge of genomes and genome structure, and the ability to solve problems independently.
Analyze genetic and genomic data that addresses fundamental questions such as, “why are individuals within species so genetically diverse, and what genetic, ecological and evolutionary processes promote that diversity?” Student is expected to have experience with linux command line and shell scripting, at least one coding language such as C/C++, Python, or R, basic knowledge of genomes and genome structure, and the ability to solve problems independently. The world’s 3 billion biodiversity specimens (insects on pins, fish in jars, fossils in drawers, plants on sheets) at museums, universities, and field stations are the fundamental source of information for reconstructing life’s historical baseline of what was where when. Biodiversity informatics is the key that scientists, natural resource managers, policymakers, and others use to unlock the potential of those specimens for addressing climate change, food scarcity, the extinction crisis, and dangers to human health. Biodiversity informatics is the creation, management, integration, analysis, and visualization of digital information related to the world’s 8 million+ species, especially as they are represented by specimens. Research topics include such things as crowdsourcing, anomaly detection, and species distribution modeling.
The world’s 3 billion biodiversity specimens (insects on pins, fish in jars, fossils in drawers, plants on sheets) at museums, universities, and field stations are the fundamental source of information for reconstructing life’s historical baseline of what was where when. Biodiversity informatics is the key that scientists, natural resource managers, policymakers, and others use to unlock the potential of those specimens for addressing climate change, food scarcity, the extinction crisis, and dangers to human health. Biodiversity informatics is the creation, management, integration, analysis, and visualization of digital information related to the world’s 8 million+ species, especially as they are represented by specimens. Research topics include such things as crowdsourcing, anomaly detection, and species distribution modeling. The McGinnis Lab studies epigenetic gene regulation in maize. Computational biology students would assist with our research by participating in analysis of RNA-seq, ChIP-seq and other large datasets. Recent and ongoing projects include co-expression analysis, identification of regulatory RNAs, and relating expression changes with epigenetic data. Students should have a basic knowledge of molecular biology and an independent working knowledge of informatics and programming (R and Python) tools and approaches.
The McGinnis Lab studies epigenetic gene regulation in maize. Computational biology students would assist with our research by participating in analysis of RNA-seq, ChIP-seq and other large datasets. Recent and ongoing projects include co-expression analysis, identification of regulatory RNAs, and relating expression changes with epigenetic data. Students should have a basic knowledge of molecular biology and an independent working knowledge of informatics and programming (R and Python) tools and approaches. Venom Genomics and Evolution: Students will analyze genomic, transcriptomic, and proteomic data to characterize venom composition and the genetic basis of adaptation in venoms of snakes, scorpions, and centipedes. Students will be expected to be comfortable working at a linux command line and coding in Python and be familiar with basic molecular biology and biochemistry.
Venom Genomics and Evolution: Students will analyze genomic, transcriptomic, and proteomic data to characterize venom composition and the genetic basis of adaptation in venoms of snakes, scorpions, and centipedes. Students will be expected to be comfortable working at a linux command line and coding in Python and be familiar with basic molecular biology and biochemistry.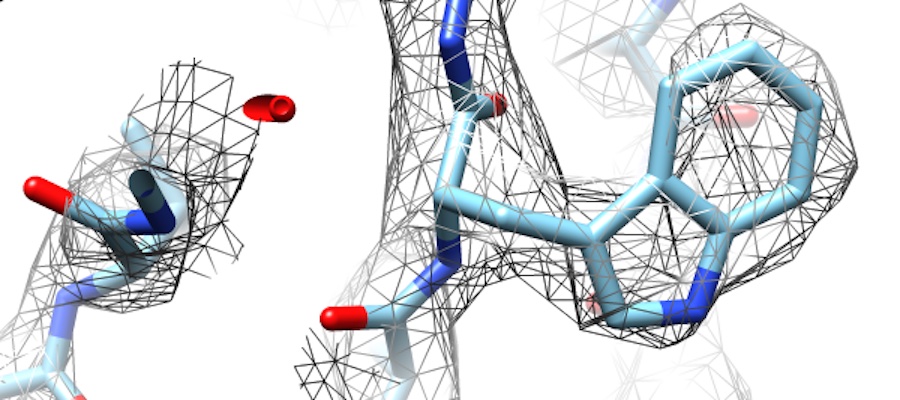 Help develop tools for high-throughput high-resolution 3D structure determination of medically relevant biomolecules. Student will be expected to have experience with linux, Python, or development of web based software.
Help develop tools for high-throughput high-resolution 3D structure determination of medically relevant biomolecules. Student will be expected to have experience with linux, Python, or development of web based software.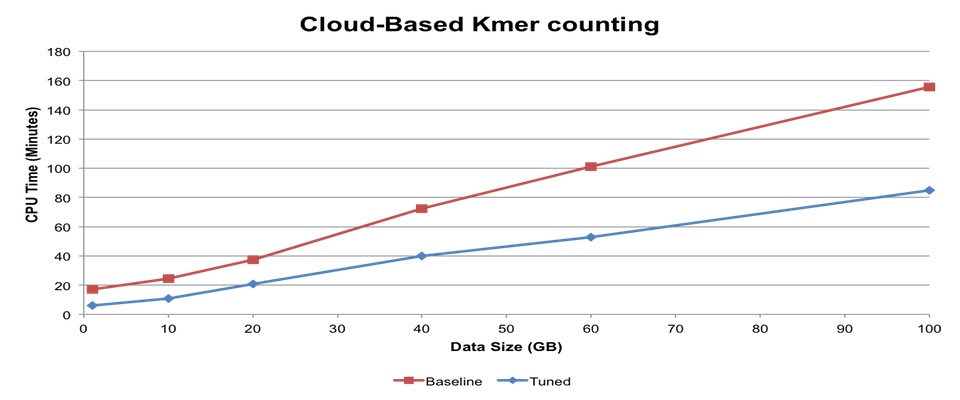 Dr. Weikuan Yu has an interest in high-performance computational biology research. In prior research projects, he has investigated various approaches to improve the performance of computational mass finger-printing, cloud-based fast k-mer counting for next-generation sequencing (NGS) data analysis and evaluation of NGS tools for meta-genomics.
Dr. Weikuan Yu has an interest in high-performance computational biology research. In prior research projects, he has investigated various approaches to improve the performance of computational mass finger-printing, cloud-based fast k-mer counting for next-generation sequencing (NGS) data analysis and evaluation of NGS tools for meta-genomics.