Hank W. Bass, Ph.D.
Research Interests
and Current Projects
SUMMARY
We are investigating the structure and function of meiotic chromosomes in the model system maize (Zea mays).Meiotic chromosome structure: Structural analysis of meiotic chromosomes invovles the creation of a new cytogenetic map of the 10 chromosomes that make up the maize genome.
Meiotic telomere function: Proper chromosome segregation during meiosis depends on the synapsis and crossing over of homologous chromosomes. Telomere movements that are unique to meiotic prophase appear to play a role in these essential homolog interactions. We are investigating the mechanism and the functional significance of meiotic telomere behavior.
METHODOLOGY
3D FISH with meiocytes.We are using 3-D fluorescence in situ hybridization (FISH) to visualize the progression of homologous chromosome pairing, chromatin remodeling, and telomere dynamics from premeiotic interphase through the end of meiotic prophase. Most of our work makes use of maize male meiotic cells that have been fixed at vairous time points throughout prophase of meiosis I.

Most of the images from these pages are single optical sections or projections of multiple sections taken from a 3-D datastack that has undergone a 3-D deconvolution to return out of focus light to it's original point sources. 3-D deconvolution is a de-blurring process that produces high quality epifluorescence imaging, near the resolution limit for fluorescence microscopy. The collection and proper redistribution of all of the photon counts collected by non-pinhole widefield imaging allows for recovery of a high level of information while resorting to relatively low levels of illumination. This allows for the repeated exposures that are needed to oversample along the Z axis, and to collect prolonged time-lapse images of living cells. This 3-D deconvolution system, developed by Sedat and Agard (UCSF), has been made commercially available by Applied Precision (API) with a system called DeltaVision.
PRIMARY PROJECTS:
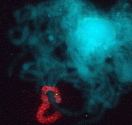
(1) Cytogenetic Map of the Maize Pachytene Karyotype
Project Website:
www.cytomaize.org
We have recently started a structural genomics project to characterize the organization of genes along the 10 chromosomes of maize. The project takes advantage alien chromosome addition lines of oat that carry single maize chromosomes (as shown at right, the maize chromosome number 9 is fluorescently stained red by FISH). Our technology provides a means of optical isolation of individual maize chromosomes. We use genomic in situ hybridization followed by 3D deconvolution microscopy. Computerize image analysis provides map position data for genes, using chromosome modeling and straightening software. FISH probes from genetically mapped sequences provide point to point links between the geneitc and the cygenetic maps. These loci are also anchored to the growing physical map from the genome sequencing project. The physical map would provide inroads for genome sequencing and map-based cloning, both important for maize, a major crop species and model system for general genetics and plant biology.
FISH Mapping Project Support
- FSU Research Foundation Cornerstone Program Enhancement Grant (PEG 1999-2001).
- Consrotium for Plant Biotechnology Research, Inc. (CPBR 2001-2002)
- National Science Foundation Plant Genome Research Program ( NSF PGRP 2003-2007)
- Bass HW, et al. (2000) Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. Journal Cell Science 113:1033-1042.
- Bass HW and Bordoli SJ. (2001) Variable distribution of meiotic homologs; on-line spinning projections of 3D data from chromosome painting and telomere FISH analysis of OMAd9.2. Maize Genetics Newsletter 75:63.
- Koumbaris G and Bass HW. (2002) Pachytene arm ratios for maize chromosome 9 in OMAd9.2, a maize chromosome addition line of oat. Maize Genetics Newsletter 76:62-63.
- Koumbaris GL and Bass HW. (2003) A new single-locus cytogenetic mapping system for maize (Zea mays L.): overcoming FISH detection limits with marker-selected sorghum (S. propinquum L.) BACs clones. The Plant Journal 35(5):647-659.
- Bassie YR, Onokpise OU, Odland WE, and Bass HW. (2004) FISH analysis of retroelement distribution patterns along mitotic chromosomes. Maize Genetics Newsletter 78:(in press)
- Anderson LK, Salameh N, Bass HW, Harper LC, Cande WZ, Weber G, and Stack SM. (2004) Integrating genetic linkage maps with pachytene chromosome structure in maize. Genetics 166:1923-1933.
- All Pubs with Links
We are interested in the remarkable and dramatic telomere rearrangements
that occur during meiotic prophase when homologous chromosomes are
undergoing pairing, synapsis, and recombination.
This work involves use of 3D molecular cytology
and molecular genetics to analyze the meiosis-specific changes in
chromatin structure and nuclear organization. Meiotic telomere clustering
is conserved in nature and results in the reorganization of the nucleus
into a structure called the bouquet, associated with homologous chromosome
synapsis. Precise control of synapsis and recombination is required for
meiotic chromosome disjunction. We wish to determine the role of
telomeres in these processes. These studies address the basic mechanisms
that underlie meiotic chromosome segregation. Thus our findings are
relevant to understanding both plant and animal genetic processes,
including those responsible for some chromosomal birth defects in
humans, such as the trisomy 21 that causes Down syndrome.
Using maize as a model system, we have identified meiotic mutants that
show altered telomere distributions
for further detailed analysis. In these
studies, a deconvolution microscope workstation in the Bass lab is
used to analyze 3D images collected from pollen mother cells obtained
year-round in FSU's Mission Road greenhouses and summer field stations.
In addition to the identification of telomere-clustering mutants, we
are analyzing telomere-binding protein genes that may be
involved in the telomere movements and nuclear architecture unique
to meiotic prophase.
Maize Telomere Research Support
- FSU Research Foundation Cornerstone Program Enhancement Grant (PEG 1999-2001).
- National Science Foundation (MCB 2001-2005)
- Bass HW, et al. (2000) Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. Journal Cell Science 113:1033-1042.
- Franklin AE, Golubovskaya I, Bass HW, and Cande WZ. (2003) Improper chromosome synapsis is associated with elongated Rad51 structures in the maize desynaptic2 mutant. Chromosoma 112:17-25.
- Bass HW, Bordoli SJ, and Foss EM. (2003) The desynaptic (dy) and desynaptic1 (dsy1) mutations in maize (Zea mays L.) cause distinct telomere-misplacement phenotypes during meiotic prophase. Journal of Experimental Botany 54(380):39-46.
- Bass HW. (2003) Telomere dynamics unique to meiotic prophase: formation and significance of the bouquet. Cellular and Molecular Life Sciences (CMLS) 60:2319-2324.
- Marian CO, Bordoli SJ, Goltz M, Santarella RA, Jackson LP, Danilevskaya O, Beckstette M, Meeley R, and Bass HW. The maize Single myb histone 1 gene, Smh1, belongs to i a novel gene family and encodes a protein that binds telomere DNA repeats in vitro. Plant Physiology 133:1336-1350.
- All Pubs with Links
MAIZE MEIOTIC CHROMOSOME IMAGES
Meiotic Prophase I DAPI Image Composites.


 CLICKAGE
stages.jpg, Annotated. 96K
CLICKAGE
stages.jpg, Annotated. 96K


more images here
ASTHETICS OF RESEARCH
 Wheel-O-Meiosis
Wheel-O-Meiosis The WHEEL-O-MEIOSIS - prophase I. It's a large jpeg composite. Some of these nuclei are DAPI stained only (courtesy RK Dawe) & others are DAPI stained after in situ hybridization (HW Bass). The bouquet structure is not evident by DAPI images alone.

Maize chromosomes have knobs, blocks of heterochromatin
made up of tandem repeats (the knob 180 repeat or TR1 sequences).
FISH-stained knobs can reveal bizzare images - especially late
at night.
Here's an example from a control experiment. The image shows a
pseudo-colored 2-wavelength overlay of DAPI (DNA-red) and FITC
(knob probe-green). This image is the raw data (pre-deconvolution),
from J. D. Lieb, inbred line W23+.
On to Images
On to Linkage
On to Lab Protocols
On to Bass Lab Home Page
HOME