Research
Early in my career, Richard
Lewontin’s book The Genetic Basis of
Evolutionary Change
impressed me. In
it, he introduced the
concept of the Genotype-Phenotype map (GP map) – the  web of
causal pathways
whereby genetic variation affects the phenotype.
This struck me as dorm-room discussion cool,
but not science practical, so I thought little about it for many years.
Since then, I made
myself first into a
population geneticist, and then into a quantitative geneticist-the two
extremes
in evolutionary genetics.
Population
genetics treats genetic variation explicitly, but then assumes (or
estimates)
that we know something about fitness effects of that variation. Quantitative genetics
incorporates the study
of phenotypic variation that causes variation in fitness, but remains
vague about
the actual genetic variation that underlies phenotypic variation. The two
disciplines remain stubbornly
independent, as they effectively use incommensurate languages about
evolution. The
means to convert between them, however, is
precisely the GP map.
web of
causal pathways
whereby genetic variation affects the phenotype.
This struck me as dorm-room discussion cool,
but not science practical, so I thought little about it for many years.
Since then, I made
myself first into a
population geneticist, and then into a quantitative geneticist-the two
extremes
in evolutionary genetics.
Population
genetics treats genetic variation explicitly, but then assumes (or
estimates)
that we know something about fitness effects of that variation. Quantitative genetics
incorporates the study
of phenotypic variation that causes variation in fitness, but remains
vague about
the actual genetic variation that underlies phenotypic variation. The two
disciplines remain stubbornly
independent, as they effectively use incommensurate languages about
evolution. The
means to convert between them, however, is
precisely the GP map.
My research program is now aimed at
actually estimating the
GP map that connects genetic and genomic variation with phenotypic
variation in
the Drosophila wing. What
once seemed
impractical now seems necessary. We
can
sequence genomes, but cannot readily determine which variants are
affecting
fitness, and more important how they might affect it.
With a GP map, we could predict what genetic
variation matters, and how phenotypes might respond to selection. Without it, we
are limited to retrospective
and correlative statements about evolution.
Predictive ability is the central challenge of science: evolutionary biologists need
to rise to that
challenge, not ignore or dismiss it.
Work in my lab uses many techniques in pursuit of this goal:
| Artificial selection experiments (graduate student Jason Cassara, post-doc Geir Bolstad). | 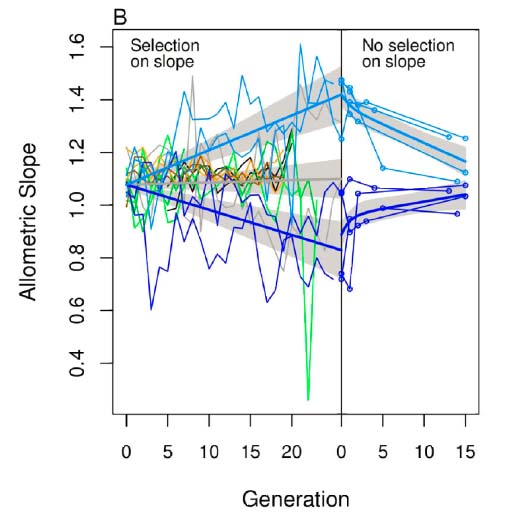 |
| Manipulations of gene expression (post-docs Eladio Marquez, and Rosa Moscarella. | 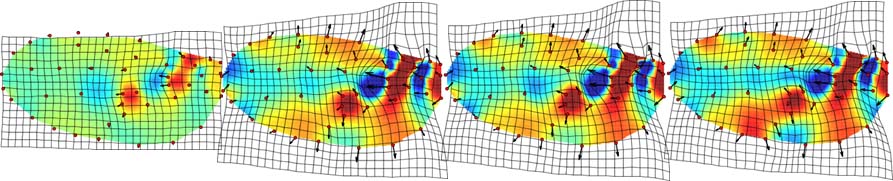 |
| Developmental morphometrics (graduate student David Aponte). | 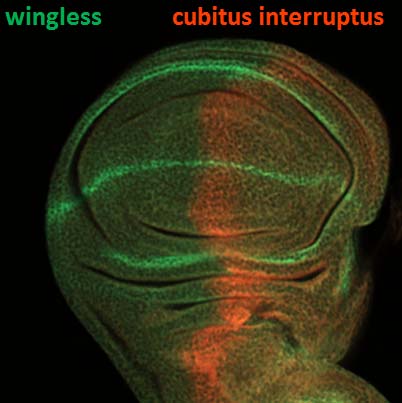 |
| Developmental modeling (post-doctoral researcher Alexis Matamoro-Vidal). | 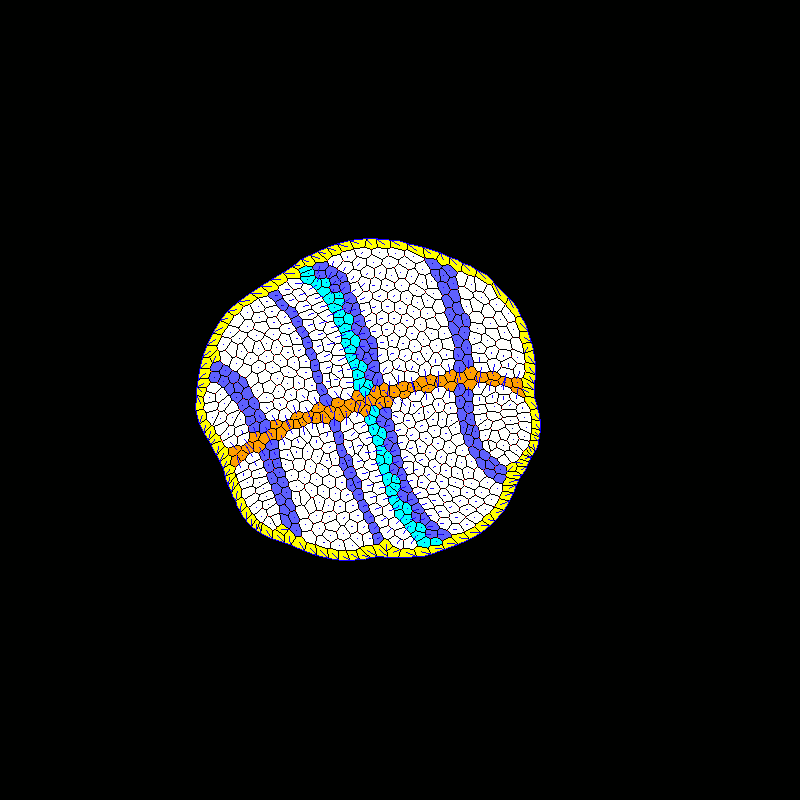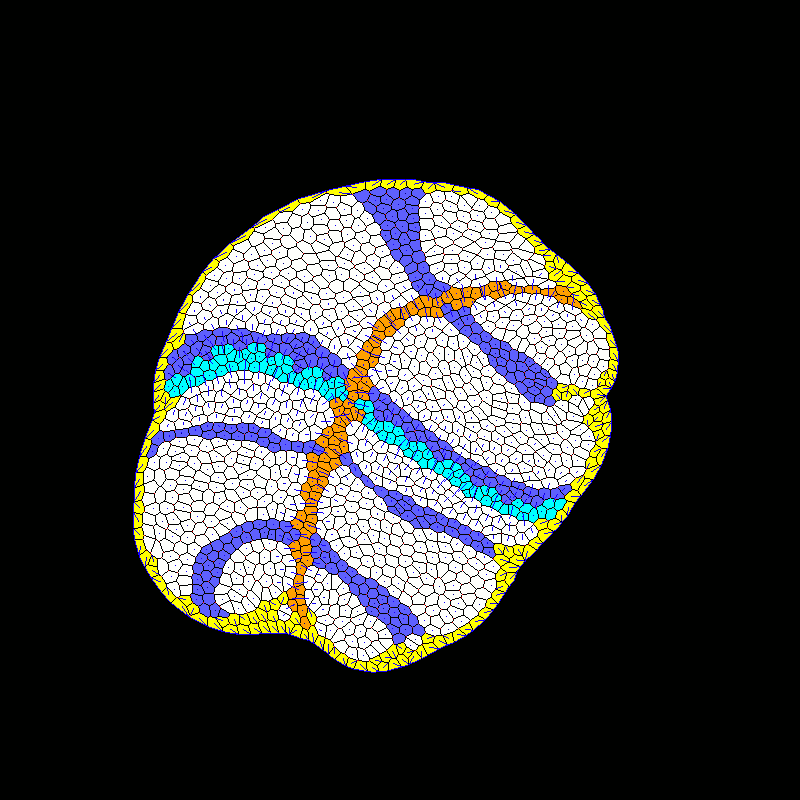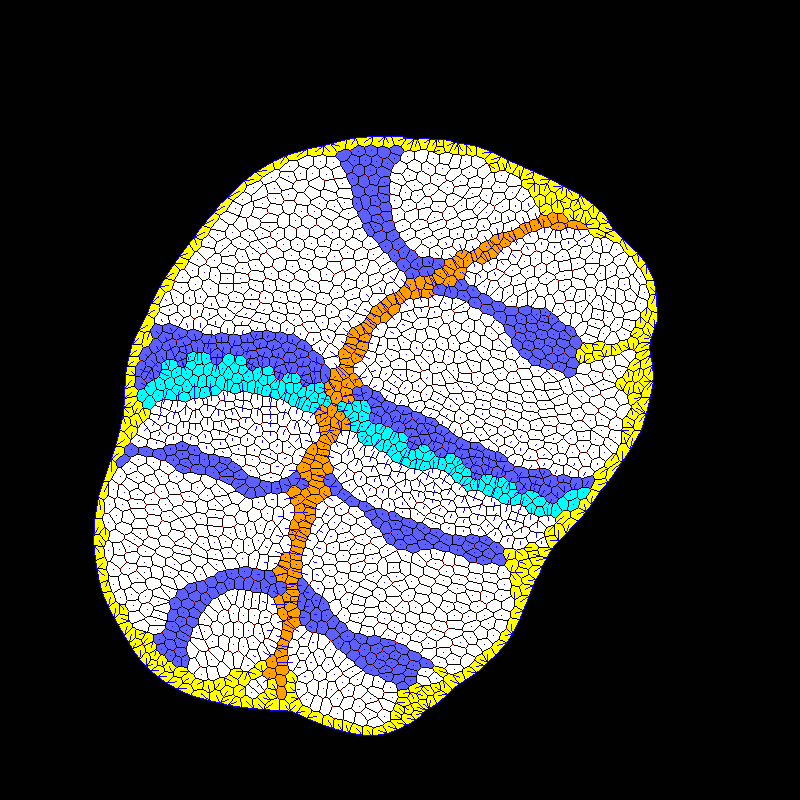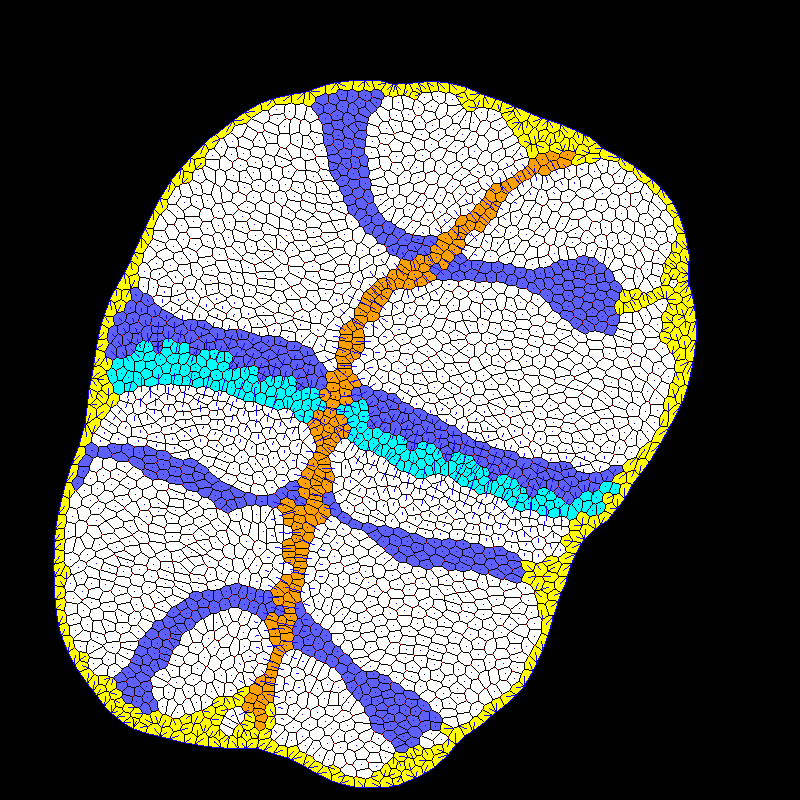 |
| Genome-wide association studies | 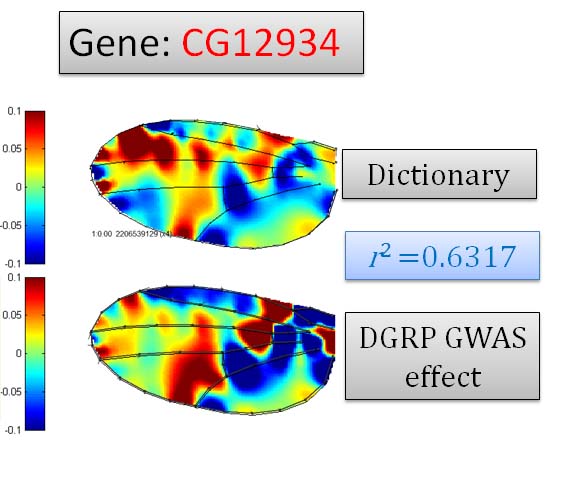 |