Position: Graduate Student
Contact: sloan@bio.fsu.edu
Hometown: Miami, FL
Education: Bachelor of Science in Biology (2010) Florida State University, Tallahassee, FL
Current Project: Genetic mapping of transgene reactivated mutant 1 (tgr1), a novel allele of the largest subunit of RNA Polymerase IV in maize.
Transcriptional gene silencing often arises through the RNA-directed DNA methylation (RdDM) pathway that results in epigenetic changes to the DNA and histones. To further understand the relationship between epigenetic modifications and gene silencing, stably silent transgenic lines containing the b1 genomic transgene (BTG-s) were used in a forward genetic screen to identify transgene reactivated (Tgr) mutants. In the M2 generation, recessive homozygous mutants were identified by purple pigmentation, characteristic of BTG expression. In addition to the loss of transcriptional silencing of BTG-s, tgr1-1 individuals exhibit hypomethylation of the promoter region of BTG-s and a reduction in 24 nucleotide siRNAs. These molecular phenotypes are consistent with Tgr1 encoding a component of the RdDM gene-silencing pathway in maize. Genetic mapping of Tgr1 indicates that the mutation lies within a 12Mbp interval on Chromosome 1, which also includes the Rmr6 locus. Complementation assays were used to demonstrate that tgr1-1 is an allele of Rmr6. We confirmed that tgr1-1 plants do not contain any of the cloned rmr6 mutant alleles. Together, these results demonstrate that tgr1-1 is a novel allele of Rmr6, a gene that has previously been shown to encode the largest subunit of RNA polymerase IV. These genetic results expand the role of Rmr6, and thus Pol IV to include the transcriptional silencing of introduced transgenes.
Previous Project: Mapping of the b1 Genomic Transgene within the Maize Genome
As an undergraduate in the lab, my independent research project focused on determining the location of a silent form of the b1 genomic transgene (Btg-s).
Importance: Identifying the location of the transgene will allow us to study the flanking sequences and their level of expression to determine the possible cause of silencing of the b1 genomic transgene.

An altered form of the b1 gene, the b1 genomic transgene (Btg), was biolistically inserted into one genotype of the maize (Zea mays) genome, thus resulting in random integration of the transgene. A silenced form of this transgene (Btg-s) is used by our lab as a marker of epigenetic regulation.
Linkage mapping is being used to isolate the location of the silent transgene. A mapping population was created by out-crossing transformants with a second genotype (B73), introducing polymorphisms into the genome that can be characterized via PCR.
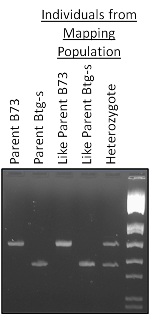 Project Flow: Allelic polymorphisms between the two genotypes (Btg-s and B73) can be analyzed through PCR amplification of various markers with known locations along each chromosome. Comparing each individual from the mapping population to both “parent” genomes allows for classification of each individuals genotype as “like B73”, “like Btg-s”, or a heterozygote of the two parents.
Project Flow: Allelic polymorphisms between the two genotypes (Btg-s and B73) can be analyzed through PCR amplification of various markers with known locations along each chromosome. Comparing each individual from the mapping population to both “parent” genomes allows for classification of each individuals genotype as “like B73”, “like Btg-s”, or a heterozygote of the two parents.
From this data allele frequencies are calculated. Random assortment predicts an allele frequency of 50% among every allele in the population. Thus choosing a population based on the presence or absence of the transgene will cause one parent to be over represented where the transgene is likely located. An allele frequency of greater than 50% corresponds to the possible location of the Btg-s.







